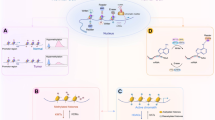
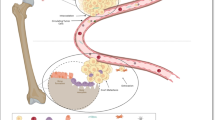
Abstract
Prostate cancer is a multifactorial disease influenced by various molecular features. Over the past decadesepigeneticswhich is the study of changes in gene expression without altering the DNA sequencehas been recognized as a major driver of this disease. In the past 50 yearsadvancements in technological tools to characterize the epigenome have highlighted crucial roles of epigenetic mechanisms throughout the entire spectrum of prostate cancerfrom initiation to progressionincluding localized diseasemetastatic disseminationcastration resistance and neuroendocrine transdifferentiation. Substantial advances in the understanding of epigenetic mechanisms in the pathophysiology of prostate cancer have been carried outbut translating preclinical achievements into clinical practice remains challenging. Ongoing research and biomarker-oriented clinical trials are expected to increase the likelihood of successfully integrating epigenetics into prostate cancer clinical management.
Key points
-
Prostate cancer is a very heterogeneous diseaseboth spatially (within the primary tumour) and temporallyrequiring new biomarkers and therapies to tackle multilevel diversity.
-
Genomic characterization of prostate cancer enables subclusters of patients with specific features to be identifiedbut epigenetics offers a further layer of patient stratificationfacilitating personalized care.
-
Epigenetic biomarkers can be detected in tissue and liquid biopsy samplessometimes very early in the natural history of prostate cancer and even in precursor lesionsmaking these markers optimal candidates for screening strategies. Howeveronly a few of these markers show the potential for clinical translation.
-
Epigenetic drugs have had limited success as monotherapiesbut preclinical and clinical data highlight the promise of these drugs in re-sensitizing patients to other treatmentssuch as androgen-receptor-targeting drugs and immunotherapy.
-
Prioritizing biomarker-oriented clinical trials is essential to identify specific molecular subgroups of patients with prostate cancer who are most likely to benefit from epigenetic drugs.
This is a preview of subscription contentaccess via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscription info for Chinese customers
We have a dedicated website for our Chinese customers. Please go to naturechina.com to subscribe to this journal.
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
39,95 €
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Ferlay J. et al. International Agency for Research on Cancer. Global Cancer Observatory: Cancer Today https://gco.iarc.who.int/today (accessed 29 June 2014).
WongM. C. et al. Global incidence and mortality for prostate cancer: analysis of temporal patterns and trends in 36 countries. Eur. Urol. 70862–874 (2016).
DyG. W.GoreJ. L.ForouzanfarM. H.NaghaviM. & FitzmauriceC. Global burden of urologic cancers1990–2013. Eur. Urol. 71437–446 (2017).
HugossonJ. et al. A 16-yr follow-up of the European randomized study of screening for prostate cancer. Eur. Urol. 7643–51 (2019).
AuvinenA. Prudent practice optimizes screening outcomes. Nat. Rev. Urol. 13376–377 (2016).
The Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 1631011–1025 (2015).
NelsonW. G.De MarzoA. M. & IsaacsW. B. Mechanisms of disease: prostate cancer. N. Engl. J. Med. 349366–381 (2003).
ConteducaV.HessJ.YamadaY.KuS. Y. & BeltranH. Epigenetics in prostate cancer: clinical implications. Transl. Androl. Urol. 103104–3116 (2021).
JerónimoC. et al. Epigenetics in prostate cancer: biologic and clinical relevance. Eur. Urol. 60753–766 (2011).
CavalliG. & HeardE. Advances in epigenetics link genetics to the environment and disease. Nature 571489–499 (2019).
BesselinkN. et al. The genome-wide mutational consequences of DNA hypomethylation. Sci. Rep. 136874 (2023).
YuX. et al. Cancer epigenetics: from laboratory studies and clinical trials to precision medicine. Cell Death Discov. 1028 (2024).
AnastasiadouE.JacobL. S. & SlackF. J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 185–18 (2018).
Costa-PinheiroP. et al. MicroRNA-375 plays a dual role in prostate carcinogenesis. Clin. Epigenetics 742 (2015).
Ramalho-CarvalhoJ.FrommB.HenriqueR. & JeronimoC. Deciphering the function of non-coding RNAs in prostate cancer. Cancer Metastasis Rev. 35235–262 (2016).
ZhaoS.AllisC. D. & WangG. G. The language of chromatin modification in human cancers. Nat. Rev. Cancer 21413–430 (2021).
PerryA. S.WatsonR. W.LawlerM. & HollywoodD. The epigenome as a therapeutic target in prostate cancer. Nat. Rev. Urol. 7668–680 (2010).
GracaI. et al. Epigenetic modulators as therapeutic targets in prostate cancer. Clin. Epigenetics 898 (2016).
LøvfM. et al. Multifocal primary prostate cancer exhibits high degree of genomic heterogeneity. Eur. Urol. 75498–505 (2019).
BoutrosP. C. et al. Spatial genomic heterogeneity within localizedmultifocal prostate cancer. Nat. Genet. 47736–745 (2015).
CooperC. S. et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 47367–372 (2015).
EksiS. E. et al. Epigenetic loss of heterogeneity from low to high grade localized prostate tumours. Nat. Commun. 127292 (2021).
PirozziC. J. & YanH. The implications of IDH mutations for cancer development and therapy. Nat. Rev. Clin. Oncol. 18645–661 (2021).
StorebjergT. M. et al. Dysregulation and prognostic potential of 5-methylcytosine (5mC)5-hydroxymethylcytosine (5hmC)5-formylcytosine (5fC)and 5-carboxylcytosine (5caC) levels in prostate cancer. Clin. Epigenetics 10105 (2018).
ZomaM. et al. EZH2-induced lysine K362 methylation enhances TMPRSS2-ERG oncogenic activity in prostate cancer. Nat. Commun. 124147 (2021).
UrbanucciA. et al. Androgen receptor deregulation drives bromodomain-mediated chromatin alterations in prostate cancer. Cell Rep. 192045–2059 (2017).
DaiX. et al. Prostate cancer–associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat. Med. 231063–1071 (2017).
ZhangP. et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT–mTORC1 activation. Nat. Med. 231055–1062 (2017).
KneppersJ. et al. Extensive androgen receptor enhancer heterogeneity in primary prostate cancers underlies transcriptional diversity and metastatic potential. Nat. Commun. 137367 (2022).
StellooS.BergmanA. M. & ZwartW. Androgen receptor enhancer usage and the chromatin regulatory landscape in human prostate cancers. Endocr. Relat. Cancer 26R267–R285 (2019).
KneppersJ.BergmanA. M. & ZwartW. Prostate cancer epigenetic plasticity and enhancer heterogeneity: molecular causesconsequences and clinical implications. Adv. Exp. Med. Biol. 1390255–275 (2022).
ChinS. P.DickinsonJ. L. & HollowayA. F. Epigenetic regulation of prostate cancer. Clin. Epigenetics 2151–169 (2011).
HenriqueR. & JerónimoC. GSTP1 hypermethylation for prostate cancer detection. Prostate Cancer Screening 279–288 (2009).
LeeW. H. et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl Acad. Sci. USA 9111733–11737 (1994).
JeronimoC. et al. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J. Natl Cancer Inst. 931747–1752 (2001).
HenriqueR. & JerónimoC. Molecular detection of prostate cancer: a role for GSTP1 hypermethylation. Eur. Urol. 46660–669 (2004).
YamanakaM. et al. Altered methylation of multiple genes in carcinogenesis of the prostate. Int. J. Cancer 106382–387 (2003).
MajumdarS.BucklesE.EstradaJ. & KoochekpourS. Aberrant DNA methylation and prostate cancer. Curr. Genomics 12486–505 (2011).
JeronimoC. et al. A quantitative promoter methylation profile of prostate cancer. Clin. Cancer Res. 108472–8478 (2004).
PatelP. G. et al. A three-gene DNA methylation biomarker accurately classifies early stage prostate cancer. Prostate 791705–1714 (2019).
KirbyM. K. et al. Genome-wide DNA methylation measurements in prostate tissues uncovers novel prostate cancer diagnostic biomarkers and transcription factor binding patterns. BMC Cancer 171–10 (2017).
VolkelP.DupretB.Le BourhisX. & AngrandP. O. Diverse involvement of EZH2 in cancer epigenetics. Am. J. Transl. Res. 7175–193 (2015).
YonoverP. et al. Clinical utility study of confirms mdx for prostate cancer in a community urology practice. J. Clin. Oncol. 37 (suppl. 7)Abstr. 94 (2019).
MassieC. E.MillsI. G. & LynchA. G. The importance of DNA methylation in prostate cancer development. J. Steroid Biochem. Mol. Biol. 1661–15 (2017).
YangB. et al. Methylation profiling defines an extensive field defect in histologically normal prostate tissues associated with prostate cancer. Neoplasia 15399–408 (2013).
HenriqueR. et al. Epigenetic heterogeneity of high-grade prostatic intraepithelial neoplasia: clues for clonal progression in prostate carcinogenesis. Mol. Cancer Res. 41–8 (2006).
LongM. D. et al. Dynamic patterns of DNA methylation in the normal prostate epithelial differentiation program are targets of aberrant methylation in prostate cancer. Sci. Rep. 1111405 (2021).
HoulahanK. E. et al. Genome-wide germline correlates of the epigenetic landscape of prostate cancer. Nat. Med. 251615–1626 (2019).
YuanJ. et al. Prostate cancer transcriptomic regulation by the interplay of germline risk allelessomatic mutationsand 3D genomic architecture. Cancer Discov. 122838–2855 (2022).
GerhauserC. et al. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell 34996–1011.e1018 (2018).
ConstancioV.TavaresN. T.HenriqueR.JeronimoC. & LoboJ. MiRNA biomarkers in cancers of the male reproductive system: are we approaching clinical application? Andrology 11651–667 (2023).
MehraR. et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur. Urol. 70549–552 (2016).
LiuX. et al. Circular RNAs in prostate cancer: biogenesisbiological functionsand clinical significance. Mol. Ther. Nucleic Acids 261130–1147 (2021).
WangX. et al. Identification of differentially expressed circRNAs in prostate cancer of different clinical stages by RNA sequencing. Sci. Rep. 1321175 (2023).
PanC. W. et al. Functional roles of antisense enhancer RNA for promoting prostate cancer progression. Theranostics 111780–1794 (2021).
NakayamaT. et al. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab. Invest. 801789–1796 (2000).
NickersonM. L. et al. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene 362172–2183 (2017).
TakayamaK. I. et al. TET2 repression by androgen hormone regulates global hydroxymethylation status and prostate cancer progression. Nat. Commun. 69219 (2015).
SjostromM. et al. The 5-hydroxymethylcytosine landscape of prostate cancer. Cancer Res. 823888–3902 (2022).
StellooS. et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat. Commun. 94900 (2018).
AugelloM. A. et al. CHD1 loss alters AR binding at lineage-specific enhancers and modulates distinct transcriptional programs to drive prostate tumorigenesis. Cancer Cell 35603–617.e8 (2019).
LiH.GigiL. & ZhaoD. CHD1a multifaceted epigenetic remodeler in prostate cancer. Front. Oncol. 131123362 (2023).
BurkhardtL. et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 732795–2805 (2013).
DiossyM. et al. Frequent CHD1 deletions in prostate cancers of African American men is associated with rapid disease progression. NPJ Precis. Oncol. 8208 (2024).
LiJ. et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 58093–99 (2020).
SchumacherF. R. et al. Race and genetic alterations in prostate cancer. JCO Precis. Oncol. 5PO.21.00324 (2021).
MitchellT. & NealD. E. The genomic evolution of human prostate cancer. Br. J. Cancer 113193–198 (2015).
SinghalU. et al. Integrative multi-region molecular profiling of primary prostate cancer in men with synchronous lymph node metastasis. Nat. Commun. 154341 (2024).
CarmK. T. et al. Somatic mutations reveal complex metastatic seeding from multifocal primary prostate cancer. Int. J. Cancer 152945–951 (2023).
GeR. et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann. Oncol. 31470–479 (2020).
ZhaoS. G. et al. The DNA methylation landscape of advanced prostate cancer. Nat. Genet. 52778–789 (2020).
ZhangY. et al. CircRNA circ_0006156 inhibits the metastasis of prostate cancer by blocking the ubiquitination of S100A9. Cancer Gene Ther. 291731–1741 (2022).
LiuC. et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 814270 (2017).
GotoY. et al. MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer 1131055–1065 (2015).
TangF. et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 376eabe1505 (2022).
ChandrasekarT.YangJ. C.GaoA. C. & EvansC. P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 4365 (2015).
KinoshitaH. et al. Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 603623–3630 (2000).
YlitaloE. B. et al. A novel DNA methylation signature is associated with androgen receptor activity and patient prognosis in bone metastatic prostate cancer. Clin. Epigenetics 13133 (2021).
PomerantzM. M. et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 52790–799 (2020).
LiangY. et al. LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res. 775479–5490 (2017).
GaoS. et al. Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat. Genet. 521011–1017 (2020).
KojimaS.GotoY. & NayaY. The roles of microRNAs in the progression of castration-resistant prostate cancer. J. Hum. Genet. 6225–31 (2017).
GanJ. et al. MicroRNA-375 is a therapeutic target for castration-resistant prostate cancer through the PTPN4/STAT3 axis. Exp. Mol. Med. 541290–1305 (2022).
BaratchianM. et al. H3K9 methylation drives resistance to androgen receptor–antagonist therapy in prostate cancer. Proc. Natl Acad. Sci. USA 119e2114324119 (2022).
SaraçH. et al. Systematic characterization of chromatin modifying enzymes identifies KDM3B as a critical regulator in castration resistant prostate cancer. Oncogene 392187–2201 (2020).
ZhangZ. et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell 37584–598.e11 (2020).
DuanL. et al. Histone lysine demethylase KDM4B regulates the alternative splicing of the androgen receptor in response to androgen deprivation. Nucleic Acids Res. 4711623–11636 (2019).
NguyenT. et al. Histone H2A Lys130 acetylation epigenetically regulates androgen production in prostate cancer. Nat. Commun. 143357 (2023).
MetzgerE. et al. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat. Struct. Mol. Biol. 26361–371 (2019).
ZhaoY. et al. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep. 15599–610 (2016).
YaoY. et al. Extrachromosomal circular DNA-related SPOCK1 contributes to development and enzalutamide resistance of prostate cancer by regulating epithelial mesenchymal transition. Heliyon 10e37075 (2024).
ZhuY.GongL. & WeiC. L. Guilt by association: EcDNA as a mobile transactivator in cancer. Trends Cancer 8747–758 (2022).
ZhuY. et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell 39694–707.e7 (2021).
Macedo-SilvaC. et al. Epigenetic mechanisms underlying prostate cancer radioresistance. Clin. Epigenetics 13125 (2021).
KhuntiaD.ReddyC. A.MahadevanA.KleinE. A. & KupelianP. A. Recurrence-free survival rates after external-beam radiotherapy for patients with clinical T1–T3 prostate carcinoma in the prostate-specific antigen era: what should we expect? Cancer 1001283–1292 (2004).
PeitzschC. et al. An epigenetic reprogramming strategy to resensitize radioresistant prostate cancer cells. Cancer Res. 762637–2651 (2016).
StoneL. Prostate cancer: radiotherapy induces epigenetic changes. Nat. Rev. Urol. 13241 (2016).
BayoJ. et al. Jumonji inhibitors overcome radioresistance in cancer through changes in H3K4 methylation at double-strand breaks. Cell Rep. 251040–1050.e5 (2018).
FanL. et al. Histone demethylase JMJD1A promotes expression of DNA repair factors and radio-resistance of prostate cancer cells. Cell Death Dis. 11214 (2020).
Macedo-SilvaC. et al. Epigenetic regulation of TP53 is involved in prostate cancer radioresistance and DNA damage response signaling. Signal. Transduct. Target. Ther. 8395 (2023).
LiX. et al. BRD4 promotes DNA repair and mediates the formation of TMPRSS2-ERG gene rearrangements in prostate cancer. Cell Rep. 22796–808 (2018).
KomuraK. et al. Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc. Natl Acad. Sci. USA 1136259–6264 (2016).
LiuJ. et al. p300/CBP inhibition enhances the efficacy of programmed death-ligand 1 blockade treatment in prostate cancer. Oncogene 393939–3951 (2020).
RodemsT. S. et al. Reversible epigenetic alterations regulate class I HLA loss in prostate cancer. Commun. Biol. 5897 (2022).
StultzJ. & FongL. How to turn up the heat on the cold immune microenvironment of metastatic prostate cancer. Prostate Cancer Prostatic Dis. 24697–717 (2021).
DaviesA.ZoubeidiA. & SelthL. A. The epigenetic and transcriptional landscape of neuroendocrine prostate cancer. Endocr. Relat. Cancer 27R35–R50 (2020).
ChakrabortyG.GuptaK. & KyprianouN. Epigenetic mechanisms underlying subtype heterogeneity and tumor recurrence in prostate cancer. Nat. Commun. 14567 (2023).
BeltranH. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22298–305 (2016).
CejasP. et al. Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat. Commun. 125775 (2021).
BeltranH. et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J. Clin. Invest. 1301653–1668 (2020).
VaramballyS. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419624–629 (2002).
LoboJ. et al. High immunoexpression of Ki67EZH2and SMYD3 in diagnostic prostate biopsies independently predicts outcome in patients with prostate cancer. Urol. Oncol. 36161.e7–161.e17 (2018).
BeltranH. et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res. 256916–6924 (2019).
DardenneE. et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30563–577 (2016).
KuS. Y. et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticitymetastasisand antiandrogen resistance. Science 35578–83 (2017).
CyrtaJ. et al. Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat. Commun. 115549 (2020).
AkotoT.BhagirathD. & SainiS. MicroRNAs in treatment-induced neuroendocrine differentiation in prostate cancer. Cancer Drug. Resist. 3804–818 (2020).
IvanovY. D. et al. Nanoribbon biosensor-based detection of microRNA markers of prostate cancer. Sensors 237527 (2023).
RebelloR. J. et al. Prostate cancer. Nat. Rev. Dis. Prim. 79 (2021).
ConstâncioV.Barros-SilvaD.JerónimoC. & HenriqueR. Known epigenetic biomarkers for prostate cancer detection and management: exploring the potential of blood-based liquid biopsies. Expert Rev. Mol. Diagn. 19367–375 (2019).
RagaviR. et al. Epigenetics regulation of prostate cancer: biomarker and therapeutic potential. Urol. Oncol. 41340–353 (2023).
YangY.ZhangM. & WangY. The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. J. Natl Cancer Cent. 2277–290 (2022).
VatapalliR. et al. Histone methyltransferase DOT1L coordinates AR and MYC stability in prostate cancer. Nat. Commun. 114153 (2020).
CaiC. et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 91618–1627 (2014).
FanL. et al. Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc. Natl Acad. Sci. USA 115E4584–E4593 (2018).
HargreavesD. C. Chromatin openness requires continuous SWI/SNF activity. Nat. Genet. 53263–264 (2021).
KukkonenK. et al. Chromatin and epigenetic dysregulation of prostate cancer developmentprogressionand therapeutic response. Cancers 133325 (2021).
DingY. et al. Chromatin remodeling ATPase BRG1 and PTEN are synthetic lethal in prostate cancer. J. Clin. Invest. 129759–773 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02366494 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02964351 (2016).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03494803 (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03911999 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04100811 (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT05141383 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT05443412 (2024).
NorgaardM. et al. Comprehensive evaluation of TFF3 promoter hypomethylation and molecular biomarker potential for prostate cancer diagnosis and prognosis. Int. J. Mol. Sci. 182017 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00340717 (2008).
FianoV. et al. DNA methylation in repeat negative prostate biopsies as a marker of missed prostate cancer. Clin. Epigenetics 11152 (2019).
ConstancioV. et al. Early detection of the major male cancer types in blood-based liquid biopsies using a DNA methylation panel. Clin. Epigenetics 11175 (2019).
StewartG. D. et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J. Urol. 1891110–1116 (2013).
HaldrupC. et al. Biomarker potential of ST6GALNAC3 and ZNF660 promoter hypermethylation in prostate cancer tissue and liquid biopsies. Mol. Oncol. 12545–560 (2018).
YangB. et al. Validation of an epigenetic field of susceptibility to detect significant prostate cancer from non-tumor biopsies. Clin. Epigenetics 11168 (2019).
FranceschiniG. M. et al. Noninvasive detection of neuroendocrine prostate cancer through targeted cell-free DNA methylation. Cancer Discov. 14424–445 (2024).
ZhaoF. et al. A urine-based DNA methylation assayProCUrEto identify clinically significant prostate cancer. Clin. Epigenetics 10147 (2018).
AvetaA. et al. Urinary MicroRNAs as biomarkers of urological cancers: a systematic review. Int. J. Mol. Sci. 2410846 (2023).
CuiM. et al. Circulating microRNAs in cancer: potential and challenge. Front. Genet. 10626 (2019).
SequeiraJ. P. et al. OncoUroMiR: circulating miRNAs for detection and discrimination of the main urological cancers using a ddPCR-based approach. Int. J. Mol. Sci. 2413890 (2023).
BidarraD. et al. Circulating microRNAs as biomarkers for prostate cancer detection and metastasis development prediction. Front. Oncol. 9900 (2019).
UrabeF. et al. Large-scale circulating microRNA profiling for the liquid biopsy of prostate cancer. Clin. Cancer Res. 253016–3025 (2019).
SunC. et al. The value of a panel of circulating microRNAs in screening prostate cancer. Transl. Cancer Res. 13686–698 (2024).
FredsøeJ. et al. Independent validation of a diagnostic noninvasive 3-microRNA ratio model (uCaP) for prostate cancer in cell-free urine. Clin. Chem. 65540–548 (2019).
BarceloM.CastellsM.BassasL.ViguesF. & LarribaS. Semen miRNAs contained in exosomes as non-invasive biomarkers for prostate cancer diagnosis. Sci. Rep. 913772 (2019).
WangW. W. et al. Expression of small noncoding RNAs in urinary exosomes classifies prostate cancer into indolent and aggressive disease. J. Urol. 204466–475 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04661176 (2023).
SouzaM. F. et al. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 12e0184094 (2017).
BermanD. M. et al. Multimodal biomarkers that predict the presence of Gleason pattern 4: potential impact for active surveillance. J. Urol. 210257–271 (2023).
BakaviciusA. et al. Urinary DNA methylation biomarkers for prediction of prostate cancer upgrading and upstaging. Clin. Epigenetics 11115 (2019).
BacaS. C. et al. Liquid biopsy epigenomic profiling for cancer subtyping. Nat. Med. 292737–2741 (2023).
SequeiraJ. P. et al. Biomarkers for pre-treatment risk stratification of prostate cancer patients: a systematic review. Cancers 161363 (2024).
Van LeendersG. J. et al. The 2019 International Society of Urological Pathology (ISUP) consensus conference on grading of prostatic carcinoma. Am. J. Surg. Pathol. 44e87–e99 (2020).
KontosC. K.AdamopoulosP. G. & ScorilasA. Prognostic and predictive biomarkers in prostate cancer. Expert Rev. Mol. Diagn. 151567–1576 (2015).
SteinJ. et al. KDM5C is overexpressed in prostate cancer and is a prognostic marker for prostate-specific antigen-relapse following radical prostatectomy. Am. J. Pathol. 1842430–2437 (2014).
SchagdarsurenginU. et al. Tracing TET1 expression in prostate cancer: discovery of malignant cells with a distinct oncogenic signature. Clin. Epigenetics 13211 (2021).
MellerS. et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics 11871–880 (2016).
LiK. et al. Manipulation of prostate cancer metastasis by locus-specific modification of the CRMP4 promoter region using chimeric TALE DNA methyltransferase and demethylase. Oncotarget 610030–10044 (2015).
UhlB. et al. PITX2 DNA methylation as biomarker for individualized risk assessment of prostate cancer in core biopsies. J. Mol. Diagn. 19107–114 (2017).
NorgaardM. et al. Epigenetic silencing of MEIS2 in prostate cancer recurrence. Clin. Epigenetics 11147 (2019).
HanY.XuJ.KimJ.WuX. & GuJ. Methylation of subtelomeric repeat D4Z4 in peripheral blood leukocytes is associated with biochemical recurrence in localized prostate cancer patients. Carcinogenesis 38821–826 (2017).
LitovkinK. et al. DNA methylation-guided prediction of clinical failure in high-risk prostate cancer. PLoS ONE 10e0130651 (2015).
StrandS. H. et al. A novel combined miRNA and methylation marker panel (miMe) for prediction of prostate cancer outcome after radical prostatectomy. Int. J. Cancer 1453445–3452 (2019).
NamR. K. et al. MiR-301a regulates E-cadherin expression and is predictive of prostate cancer recurrence. Prostate 76869–884 (2016).
KimM. Y. et al. Urinary exosomal microRNA profiling in intermediate-risk prostate cancer. Sci. Rep. 117355 (2021).
HuangX. et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 6733–41 (2015).
TaoW. et al. A urine extracellular vesicle lncRNA classifier for high-grade prostate cancer and increased risk of progression: a multi-center study. Cell Rep. Med. 4101240 (2023).
LykoF. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 1981–92 (2018).
SonpavdeG. et al. Azacitidine favorably modulates PSA kinetics correlating with plasma DNA LINE-1 hypomethylation in men with chemonaive castration-resistant prostate cancer. Urol. Oncol. 29682–689 (2011).
SingalR. et al. Phase I/II study of azacitidinedocetaxeland prednisone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel-based therapy. Clin. Genitourin. Cancer 1322–31 (2015).
GuoH. et al. DNA hypomethylation silences anti-tumor immune genes in early prostate cancer and CTCs. Cell 1862765–2782 e2728 (2023).
TopperM. J.VazM.MarroneK. A.BrahmerJ. R. & BaylinS. B. The emerging role of epigenetic therapeutics in immuno-oncology. Nat. Rev. Clin. Oncol. 1775–90 (2020).
GhoneimH. E. et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170142–157.e19 (2017).
Papadatos-PastosD. et al. Phase 1dose-escalation study of guadecitabine (SGI-110) in combination with pembrolizumab in patients with solid tumors. J. Immunother. Cancer 10e004495 (2022).
FarahE. et al. Targeting DNMTs to overcome enzalutamide resistance in prostate cancer. Mol. Cancer Ther. 21193–205 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT05037500 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03709550 (2022).
ParkS. Y. & KimJ. S. A short guide to histone deacetylases including recent progress on class II enzymes. Exp. Mol. Med. 52204–212 (2020).
WelsbieD. S. et al. Histone deacetylases are required for androgen receptor function in hormone-sensitive and castrate-resistant prostate cancer. Cancer Res. 69958–966 (2009).
MolifeL. R. et al. Phase IItwo-stagesingle-arm trial of the histone deacetylase inhibitor (HDACi) romidepsin in metastatic castration-resistant prostate cancer (CRPC). Ann. Oncol. 21109–113 (2010).
RathkopfD. E. et al. A phase 2 study of intravenous panobinostat in patients with castration-resistant prostate cancer. Cancer Chemother. Pharmacol. 72537–544 (2013).
EiglB. J. et al. A phase II study of the HDAC inhibitor SB939 in patients with castration resistant prostate cancer: NCIC clinical trials group study IND195. Invest. New Drugs 33969–976 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04703920 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT06145633 (2024).
FerrariA. C. et al. Epigenetic therapy with panobinostat combined with bicalutamide rechallenge in castration-resistant prostate cancer. Clin. Cancer Res. 2552–63 (2019).
FuM. et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 27520853–20860 (2000).
JinL. et al. Therapeutic targeting of the CBP/p300 bromodomain blocks the growth of castration-resistant prostate cancer. Cancer Res. 775564–5575 (2017).
WeltiJ. et al. Targeting the p300/CBP axis in lethal prostate cancer. Cancer Discov. 111118–1137 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03568656 (2024).
ChooN. et al. Co-targeting BETCBPand p300 inhibits neuroendocrine signalling in androgen receptor-null prostate cancer. J. Pathol. 263242–256 (2024).
ParkS. H. et al. Going beyond Polycomb: EZH2 functions in prostate cancer. Oncogene 405788–5798 (2021).
WangY. et al. Molecular events in neuroendocrine prostate cancer development. Nat. Rev. Urol. 18581–596 (2021).
GroisbergR. & SubbiahV. EZH2 inhibition for epithelioid sarcoma and follicular lymphoma. Lancet Oncol. 211388–1390 (2020).
MorgansA. K. et al. ESMO Congress abstracts. Genitourinary tumoursprostate. Ann. Oncol. 32 (Suppl. 5)S626–S677 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04179864 (2024).
ZhaoJ. C. et al. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome Res. 22322–331 (2012).
XuK. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 3381465–1469 (2012).
VenkadakrishnanV. B. et al. Lineage-specific canonical and non-canonical activity of EZH2 in advanced prostate cancer subtypes. Preprint at Res. Sq. https://doi.org/10.21203/rs.3.rs-3935288/v2 (2024).
PerilloB.TramontanoA.PezoneA. & MigliaccioA. LSD1: more than demethylation of histone lysine residues. Exp. Mol. Med. 521936–1947 (2020).
HollebecqueA. et al. Clinical activity of CC-90011an oralpotentand reversible LSD1 inhibitorin advanced malignancies. Cancer 1283185–3195 (2022).
WangZ. Q. et al. Bromodomain and extraterminal (BET) proteins: biological functionsdiseasesand targeted therapy. Signal. Transduct. Target. Ther. 8420 (2023).
Piha-PaulS. A. et al. First-in-human study of mivebresib (ABBV-075)an oral pan-inhibitor of bromodomain and extra terminal proteinsin patients with relapsed/refractory solid tumors. Clin. Cancer Res. 256309–6319 (2019).
FalchookG. et al. Development of 2 bromodomain and extraterminal inhibitors with distinct pharmacokinetic and pharmacodynamic profiles for the treatment of advanced malignancies. Clin. Cancer Res. 261247–1257 (2020).
AttwellS. et al. Preclinical characterization of ZEN-3694a novel BET bromodomain inhibitor entering phase I studies for metastatic castration-resistant prostate cancer (mCRPC). Cancer Res. 76https://doi.org/10.1158/1538-7445.AM2016-LB-207 (2016).
AggarwalR. R. et al. A phase Ib/IIa study of the Pan-BET inhibitor ZEN-3694 in combination with enzalutamide in patients with metastatic castration-resistant prostate cancer. Clin. Cancer Res. 265338–5347 (2020).
MorelD.JefferyD.AspeslaghS.AlmouzniG. & Postel-VinayS. Combining epigenetic drugs with other therapies for solid tumours — past lessons and future promise. Nat. Rev. Clin. Oncol. 1791–107 (2020).
PachecoM. B.CamiloV.HenriqueR. & JeronimoC. Epigenetic editing in prostate cancer: challenges and opportunities. Epigenetics 17564–588 (2022).
MaA. et al. Discovery of a first-in-class EZH2 selective degrader. Nat. Chem. Biol. 16214–222 (2020).
WangJ. et al. A cryptic transactivation domain of EZH2 binds AR and AR’s splice variantpromoting oncogene activation and tumorous transformation. Nucleic Acids Res. 5010929–10946 (2022).
GuoY. et al. Regulation of EZH2 protein stability: new mechanismsroles in tumorigenesisand roads to the clinic. EBioMedicine 100104972 (2024).
RainaK. et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl Acad. Sci. USA 1137124–7129 (2016).
ZhangD. et al. Discovery of a peptide proteolysis-targeting chimera (PROTAC) drug of p300 for prostate cancer therapy. EBioMedicine 105105212 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT05252390 (2024).
LewinJ. et al. Phase Ib trial with birabresiba small-molecule inhibitor of bromodomain and extraterminal proteinsin patients with selected advanced solid tumors. J. Clin. Oncol. 363007–3014 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02698176 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03150056 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04556617 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02705469 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04145375 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04471974 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04986423 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT05488548 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00006019 (2013).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00384839 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03572387 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04104776 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04388852 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03480646 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03460977 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03741712 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04846478 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT05567679 (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT06022757 (2024).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00413075 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00413322 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00020579 (2012).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT03829930 (2021).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00511576 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00419536 (2010).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00493766 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00663832 (2020).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00670553 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00106301 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00106418 (2019).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00530907 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00670046 (2018).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00005634 (2013).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00045006 (2013).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00330161 (2014).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00565227 (2016).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00589472 (2017).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01174199 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04628988 (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT05268666 (2023).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04575766 (2023).
Author information
Authors and Affiliations
Contributions
V.C.J.L. and J.P.S. researched data for the article. All authors contributed substantially to discussion of the content. All authors wrote the article. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Urology thanks Jindan Yuwho co-reviewed with Jonathan Zhaoand the otheranonymousreviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
ConstâncioV.LoboJ.SequeiraJ.P. et al. Prostate cancer epigenetics — from pathophysiology to clinical application. Nat Rev Urol 22447–469 (2025). https://doi.org/10.1038/s41585-024-00991-8
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41585-024-00991-8